New England BioLabs (NEB)
Q5® high-fidelity DNA polymerase
The finest PCR fidelity in over 10 years!
What is fidelity or “Proofreading activity” of a Polymerase?
DNA polymerase fidelity describes the replication accuracy of a desired template during DNA amplification, also named “proofreading” activity. High-fidelity DNA polymerases have several checkpoints to protect against making and propagating mistakes while copying DNA:
1. Precision of template strand reading
2. Correctness of nucleoside triphosphates inserted
3. Replacement of incorrect nucleotides incorporated
1. The precision of template strand reading
Precision reading of the template strand means that the polymerase enzyme interprets the exact nucleoside triphosphates order exposed on the DNA strand to be replicated. This ability of the fidelity polymerases is obtained by a significant binding preference for the correct versus the incorrect nucleotide triphosphate during the DNA polymerization.
2. Correctness of nucleoside triphosphates inserted
Correctness of nucleoside triphosphates inserted is ensured by selecting and inserting the correct nucleoside triphosphates at the 3'-primer terminus in accordance with Watson-Crick base pairing. If an incorrect nucleotide does bind in the polymerase active site, incorporation is slowed due to the sub-optimal architecture of the active site complex. This time increases the opportunity for the incorrect nucleotide to dissociate before incorporation, thereby allowing the process to start again (and for a correct nucleotide triphosphate to bind) (ref. 1 and 2).
3. Replacement of incorrect nucleotides incorporated
Finally, the replacement of incorrect nucleotides incorporated is checked by effective discrimination of correct versus incorrect nucleotide incorporation. If an incorrect nucleotide is inserted, proofreading DNA polymerases have an extra line of defense.
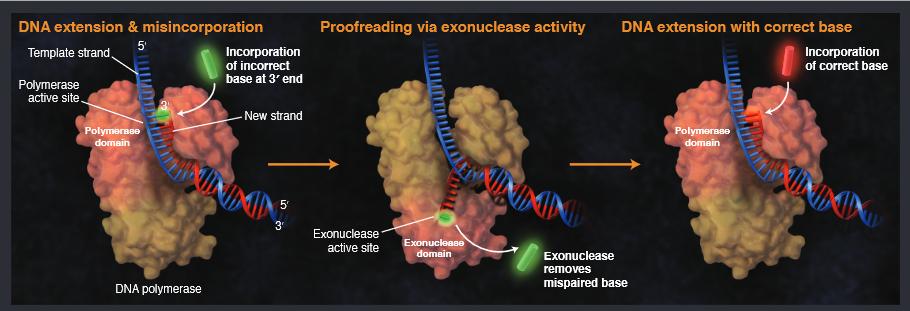
They can "sense" the perturbation caused by the mispaired bases and move the 3' end of the growing DNA chain into a proofreading 3'→5' exonuclease domain. The incorrect nucleotide is removed by the 3'→5' exonuclease activity before the chain is moved back into the polymerase domain, where polymerization can continue with the correct nucleotide (see Figure 1).
Figure 1. DNA replication with a proofreading polymerase
Extension proceeds along the template strand at the 3' end of the newly synthesized strand. When the polymerase recognizes an error, the mismatched base is transferred to the exonuclease active site, and the base is excised. The extended strand returns to the polymerase domain, re-anneals to the template strand, and replication continues.
When is fidelity important?
Fidelity is important for applications in which the DNA sequence must be correct after the amplification, including:
- Cloning/subcloning protein expression
- SNP (Single Nucleotide Polymorphism) analysis
- RNA analysis by RT-PCR
- Next generation sequencing applications
On the other hand, fidelity is less important for many diagnostic applications where the read-out is simply the presence or absence of a product.
Why is Q5 so proofreading and robust?
New England Biolabs (NEB) has been involved in the discovery and development of high-fidelity polymerases for many years. Beginning with the discovery of Vent® DNA Polymerase from Thermococcus litoralis in 1990. Since then, NEB researchers have identified and engineered several high-fidelity polymerases.
In 2012, the Q5 DNA polymerase was released by NEB as a new standard for both fidelity and robust performance. The fidelity of Q5, measured as >280 times* higher than Taq polymerase, ensures ultra-low error rates. In addition, Q5 demonstrates:
- Robust amplification with minimal optimization
- Superior coverage for a broad range of amplicons, regardless of GC content
- Shorter PCR protocols
- Amplify templates up to 20 kb
* Fidelity comparisons between polymerases can be expressed in absolute terms, often by the number of errors per 1,000 or 10,000 nucleotides or expressed as the number of theoretical errors per base. The level of fidelity can also be expressed in relative terms, often using Taq DNA polymerase as the relative standard.
Summary
The superior proofreading activity of Q5 DNA polymerase is caused by a fusion of to the processivity-enhancing Sso7d DNA binding domain polymerase from Sulfolobus solfataricus, a species of thermophilic archaeon isolated and discovered in the Solfatara volcano. This fusion improves fidelity (more than 50-fold higher than Taq), but also speed and performance reliability (amplifying targets up to 20 kb long with extension times as low as 10 seconds per kb). In addition, the Q5 buffer system has been extensively optimized to provide robust performance across both high-GC and high-AT regions, as well as other challenging amplicons.
| Cat. no. | Description | Size |
|---|---|---|
| M0491S | Q5 High-Fidelity DNA Polymerase | 100 units |
| M0491L | Q5 High-Fidelity DNA Polymerase | 500 units |
| M0493S | Q5 Hot Start High-Fidelity DNA Polymerase | 100 units |
| M0493L | Q5 Hot Start High-Fidelity DNA Polymerase | 500 units |
| M0492S | Q5 High-Fidelity 2X Master Mix | 100 reactions |
| M0492L | Q5 High-Fidelity 2X Master Mix | 500 reactions |
| M0494S | Q5 Hot Start High-Fidelity 2X Master Mix | 100 reactions |
| M0494L | Q5 Hot Start High-Fidelity 2X Master Mix | 500 reactions |
1. Johnson, K.A. (2010) Biochemica et Biophysica Acta, 1804, 1041–1048.
2. Joyce, C.M. and Benkovic, S.J. (2004) Biochemistry, 43, 14317–14324.
Request a free sample
Reach out for more information!

