Blog article
Having trouble when amplifying GC-rich sequences?
Here are four tips on how to troubleshoot
Written by our Swedish colleagues.
What is the meaning of GC-rich and why can it be challenging to amplify?
A GC-rich template refers to a DNA sequence where 60% (or greater) of the bases present are G (guanine) or C (cytosine). Only 3% of the human genome is GC-rich, but these regions are often found in the promoters of genes, particularly housekeeping and tumor suppressor genes. Three hydrogen bonds are established in a G-C base pair in contrast to the two between an A-T base pair. This renders a G-C bond more thermostable, as three hydrogen bonds require more energy to break. GC-rich regions are also ‘bendable’, readily forming secondary structures like hairpins. If you see a blank gel or a DNA smear when trying to amplify a GC-rich template, consider your reagents (polymerase choice, Mg2+ concentration, and the effects of various additives) and the annealing temperature of your reaction.
Master mix formats are ideal for convenience and reducing pipetting errors, however they bring little flexibility to the user setup when troubleshooting a GC-rich PCR. For this reason, some master mixes are specifically tailored to amplify GC-rich sequences (e.g., OneTaq® Hot Start 2X Master Mix with GC Buffer). Alternatively, if you choose to use a standalone polymerase, it is easier to tweak the reaction components when optimizing.
1. Polymerase choice
Choosing the right polymerase to amplify GC-rich regions is critical. Taq polymerase is the most common polymerase used for PCR, but now, many polymerases have been specifically optimized to amplify GC-rich sequences. Polymerases can sometimes struggle with the complex secondary structures that tend to form when GC-rich stretches fold onto themselves, resulting in a block of the enzyme and consequently synthesis of shorter/incomplete molecules. Furthermore, GC-rich regions resist denaturation, making it difficult for primers to anneal. And the primers used to amplify GC-rich templates tend to form dimers. Changing the polymerase can sometimes overcome these challenges
Tip:
Some polymerases are supplied with a GC Enhancer that contains additives that help to inhibit secondary structure formation and increase primer stringency. OneTaq DNA Polymerase (NEB #M0480) is twice the fidelity of Taq polymerase and is ideal for routine or GC-rich PCR. It was developed with both standard and GC buffers to provide high yield and specificity for particularly difficult amplicons. Up to 80% GC content can be amplified by adding the OneTaq High GC Enhancer to the GC buffer.
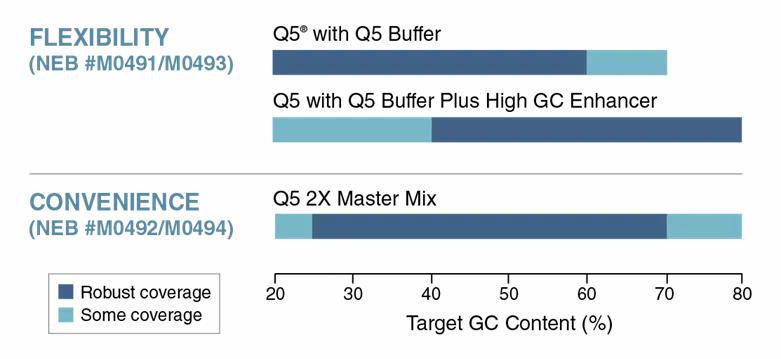
For more challenging areas, the Q5® High-Fidelity DNA Polymerase (NEB #M0491) is more than 280 times the fidelity of Taq and is ideal for long or difficult amplicons, including GC-rich DNA. Amplification can be improved on GC-rich sequences by adding Q5 High GC Enhancer to the buffer. The figure below shows the robust performance of the Q5 High-Fidelity 2X Master Mix (NEB #M0492) on 25 to 70% GC content. In comparison, the standalone polymerase provides flexibility to amplify a wider range of GC content and can give robust performance up to 80% GC, thanks to the GC enhancer supplied with the polymerase.
Q5 formats allow robust amplification of a broad range of targets.
On a side note – if you are working with blood samples, NEB has just released their Q5 Blood Direct 2X Master Mix (NEB #M0500), which works well for amplicons up to 75% GC content. The buffer delivers increased resistance to inhibitors in the blood. You can fast-track your workflow by amplifying targets directly from dried blood spots or up to 30% whole human blood, skipping the DNA purification step.
2. Mg2+ concentration
Magnesium is a cofactor in PCR. It has some major roles in the reaction, being required for the enzymatic activity of the polymerase and enabling the addition of dNTPs, besides being essential for primer binding. Too much MgCl2 can lead to non-specific primer binding during the annealing step, which will be viewed as multiple DNA bands on an agarose gel. On the other hand, too little MgCl2 will cause reduced polymerase activity leading to weak or no amplification.
MgCl2 is typically supplied in the buffer that accompanies the polymerase. For standard PCR reactions, 1.5 to 2 mM MgCl2 is the most used concentration. However, DNA templates with high GC content may require altering the concentration.
Tip:
If you suspect that the Mg2+ concentration is the cause of a failed PCR, we advise you to try a concentration gradient of MgCl2 to find that sweet spot where you can eliminate non-specific binding while still maximizing your yield. Try 0.5 mM increments between 1.0 and 4 mM.
3. Effects of additives
Many known additives impact the amplification of GC-rich regions. They work by either:
- reducing secondary structures, which increases amplification of your target, or
- reducing non-specific priming and the amplification of off-target DNA.
DMSO, Glycerol and Betaine reduce secondary structures that can inhibit the polymerase.
Formamide and Tetramethyl ammonium chloride increase the primer annealing stringency, which increases specificity.
7-deaza-2′-deoxyguanosine is a dGTP analog that can improve the PCR yield of GC-rich regions, although it can be challenging to intercalate some staining agents.
Tip:
If you don’t know the cause of your poor amplification, you might not know which of these additives will work the best. It is laborious to test them all using various concentration gradients for each. Furthermore, testing each additive individually will bring numerous new variables to your optimization workflow.
New England Biolabs® has formulated two polymerases with optimized GC Enhancers (discussed above in the polymerase section) that contain various GC-rich PCR enhancing additives: OneTaq DNA Polymerase (NEB #M0480), and Q5 High-Fidelity DNA Polymerase (NEB #M0491).
4. Annealing temperature
Multiple bands on a gel are an indication of non-specific binding. You might need to increase your annealing temperature (Ta). If you’re getting no product at all, perhaps the annealing temperature is too high. The Tm is the primer melting temperature, and Ta is the primer annealing temperature. It’s typical to design primers with a Tm of between 50 and 72°C, and the primer annealing temperature (Ta) should be about 5°C lower than the Tm. The Tm is the temperature at which there is an equilibrium between the free and template-bound primer, considering that primer is usually the excess reagent.
As mentioned above, a G-C pair has three hydrogen bonds, and an A-T pair has two hydrogen bonds. Based on this, you can predict that the higher the GC-content, the higher the temperature required to disrupt them. Generally, a higher Ta results in more specific primer binding but less product formation (so more PCR cycles might be required). But if the annealing temperature is too high, the primer will have reduced template hybridization.
Tip:
To prevent non-specific amplification, you can try a higher annealing temperature (Ta). This can also help separate secondary structures. Try a temperature gradient or use a higher annealing temperature for the first few PCR cycles.
The NEB web tool Tm Calculator can help you evaluate the best Ta for your PCR reaction; it takes into account the enzyme and buffer you intend to use.
It’s important to keep in mind that there isn’t a universal solution that works well for all GC-rich amplicons. The impact of changing any parameter outlined above will be target specific, so what works for one amplicon may not work for another (e.g., you may need 10% OneTaq GC Enhancer for one target but 20% for another). Optimization is required for each target.
This blog article is adapted from Gibson J. “Four tips for PCR amplification of GC-rich sequences”, nebinspired-blog.
https://international.neb.com/nebinspired-blog/four-tips-for-pcr-amplification-of-gc-rich-sequences
Reach out for more information!

